Research
Our main research question is to understand the genomic bases of the evolution of new body plans. To answer this , we focus on key evolutionary transitions and major genomic changes, such as the origin of vertebrates or echinoderms. We used a combination of comparative genomics and regulatory profiling techniques to trace back the origin of new genes and new regulatory mechanisms. Our long term goal is to develop computational approach to extend the comparative methods of phylogenetics and evolutionary genomics to new classes of functional genomics datasets describing active chromatin regions, chromatin interactions and cellular resolution gene expression.
Origin of echinoderm bodyplan
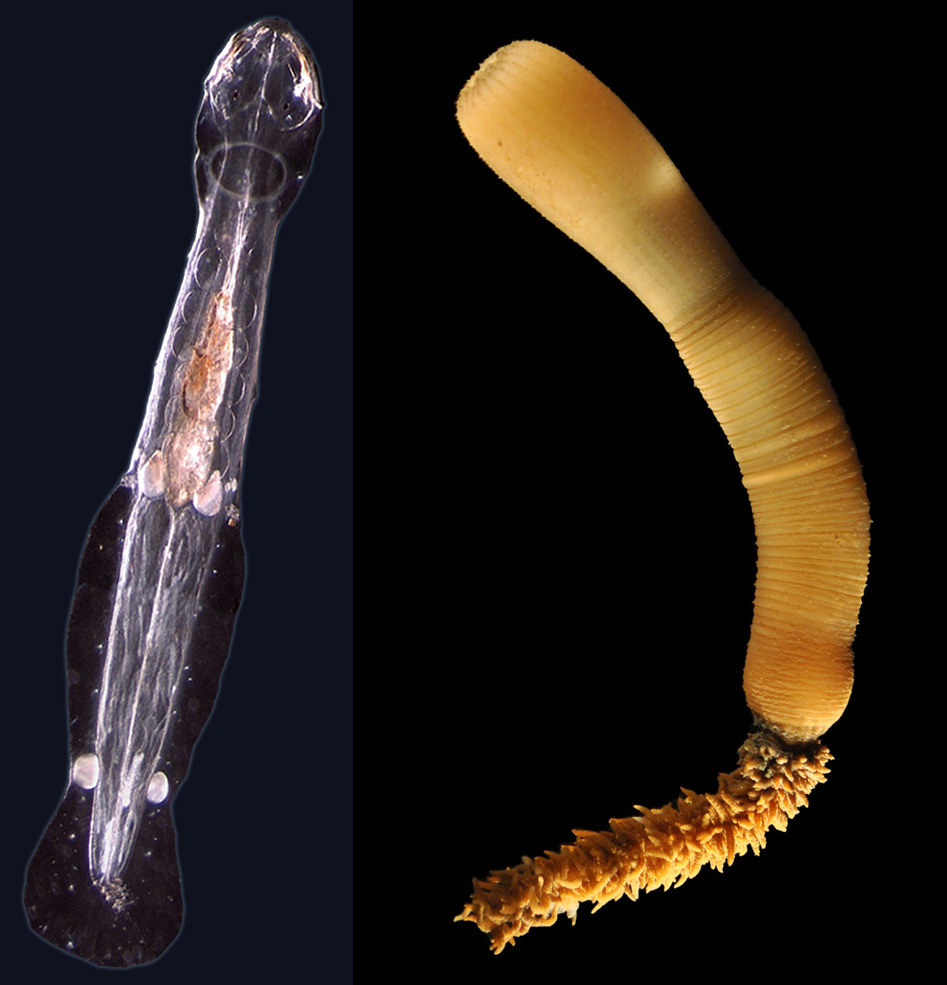
Echinoderms are the only animals to show a pentaradial star-like symmetry while they evolved from animals that have a bilateral - mirror-like - symmetry. Our goal is to understand how developmental genes have been redeployed to establish this unique bodyplan among animals. Particularly, we want to study how changes of the genome architecture have altered gene regulation and made it possible for genes to acquire new expression patterns during metamorphosis. For instance, Hox genes pattern anterioposterior symmetry in animals, but in some echinoderm lineages they have been redeployed during larval development and metamorphosis, while at the same time, their conserved genomic organisation in a tight cluster has been rearranged in some echinoderm lineages (e.g. sea urchin).
The picture on the left shows the (pentaradial) rudiment of an adult sea urchin growing within the larva during the metamorphosis process.
We apply comparative genomics to new and existing echinoderm genomes (sea urchins, sea stars, sea brittle, sea cucumbers, sea lilies) in order to trace back the genomic changes characteristics of echinoderms in terms of gene order, genome organisation and gene content. We deploy functional genomics approaches, such as ATAC-seq and HiC-seq to profile the regulatory events that take place during the metamorphosis of the echinoderm bilateral larval into a pentaradial adult. We also assess more specifically the role of functional genomic elements using transgenesis in sea urchin.
To know more about sea urchin development, check this video from planktonchronicles.org:
Genomes duplications in vertebrates
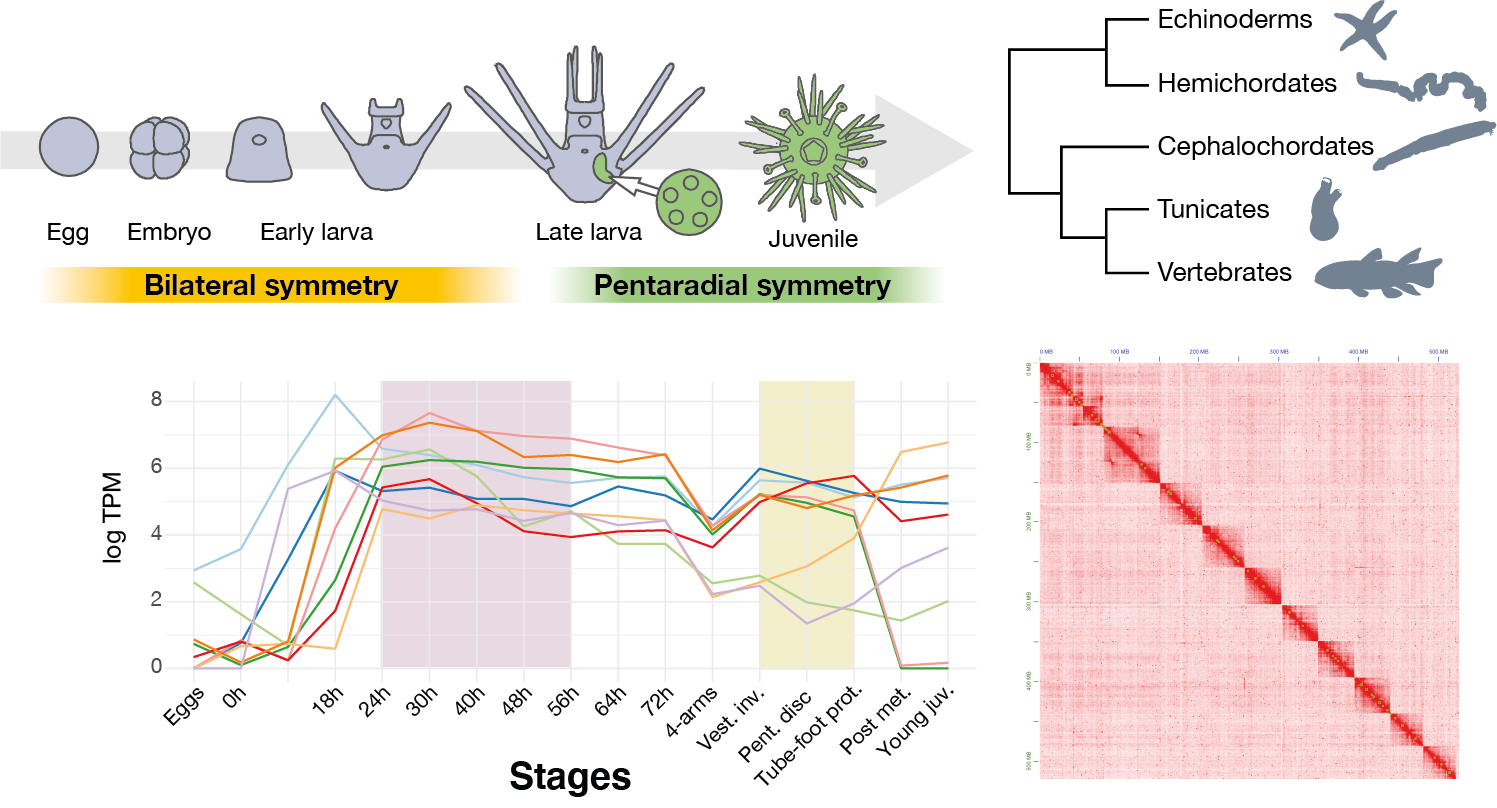
The surge of new characters in vertebrates, such as limbs, sensory organs, brain segmentation, is a key event in the history of animals, marginally because it gave rise to the human descents. Interestingly, these innovations have often been related to a drastic genomic event, -two round of whole genome duplication (WGDs) that took place at the origin of vertebrates. Amphioxus is a member of cephalochordates, the earliest chordate lineage that diverged before these whole genome duplications and studying its genome make it possible to better understand the changes that took place in vertebrates. Recent progresses in which we have participages have shows that vertebrate regulatory lanscape was affected by WGDs and showed an increased number of regulatory elements. Similarly, the first chromosome-scale genome of amphioxus has demonstrated that vertebrate chromosomes have been heavily rearranged after WGDs.
Our future work will attempt to understand the relationships between these architectural changes and the newly evolved gene regulation. It will also focus on better decipher the exact sequence and nature of WGDs and the evolution of vertebrate chromosomes. Notably, the exact history of WGDs in the earliest vertebrate lineages, the cyclostomes, remains elusive. To better understand that, we are studying the genomes and gene expression in organisms as animals as amphioxus, skate, sharks, lampreys and hagfish (see below).
Evolution and phylogeny of animal genomes
We are convinced that studying genome organisation in the broadest diversity of animal lineages could illuminate the evolution of many genome traits and help understand how bodyplan evolve. Two key questions that we are interested in are:
- The evolution of ancestral chromosomal linkage in animals. Many but not all animals have presever the ancestral chromsomal idendity among animals, we want to resolve chromosomal organisation in key animal lineages to examine the evolutionary dynamics of chromosomal organisation and determine whether they can provide phylogenetic information.
- Using complete genome to improve the phylogentic reconstruction of the tree of life. Our goal is to extend the breadth of phylogenetic characters available: genome structure, gene family evolution and genome organisation, etc…
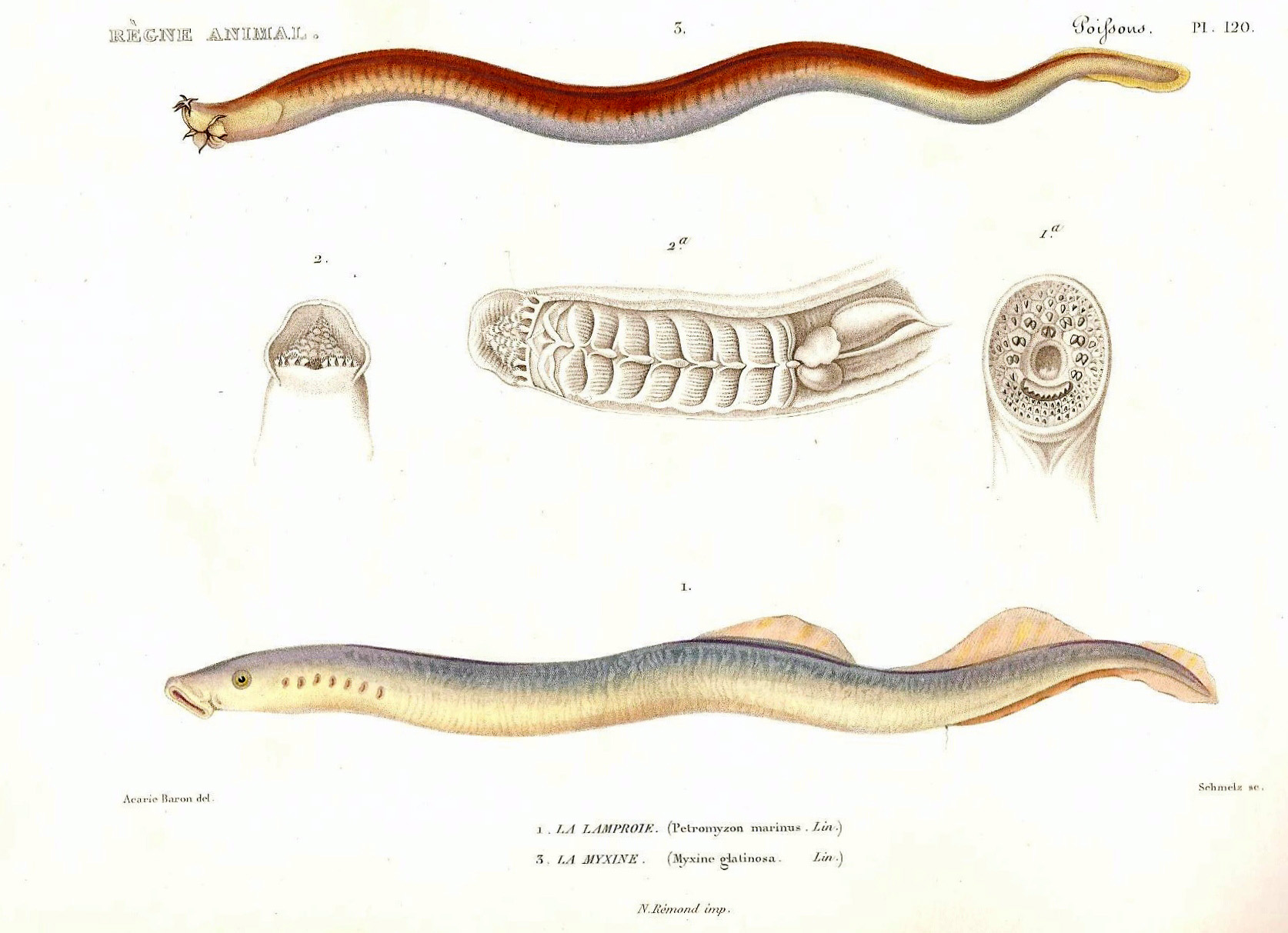
Two of our favorite upcoming genomes are the chaetognaths or arrow worms (left) and the priapulid or penis worm (right). Chaetognaths are planktonic predators whose phylogenetic position has been recently updated to the earliest lineage of the Lophotrochozoan branch (aka Spiralians) and they show a unique and highly original bodyplan. Priapulids is one of the early branching ecdysosoan lineage and studying their genome can help understand gene losses that took place this lineage. Interestingly, both lineages show deuteronomous development that could be reminiscent of the ancestral state in bilaterians.